Full Time Professor
Bioengineering and Chemical Engineering Department
CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) technology is a genetic editing technique that acts as a molecular “scissors” using an RNA molecule as a cutting guide. This tool is often used to change one or, at most, a few DNA bases. It breaks the two strands of DNA and then relies on the cell’s DNA repair systems to ligate the strands back together, introducing new mutations or deletions. However, the method exposes the DNA breaks, which can lead to undesired errors. It has been estimated that CRISPR has an error rate of around 4-5%. Although the editing is not perfect, CRISPR has revolutionized the biomedical industry and medicine by enabling direct edits to genes. As mentioned, CRISPR is well-suited for small but not for large edits. The recent discovery of a molecular editing mechanism known as “bridge RNA,” found in bacteria, could overcome CRISPR’s limitations.
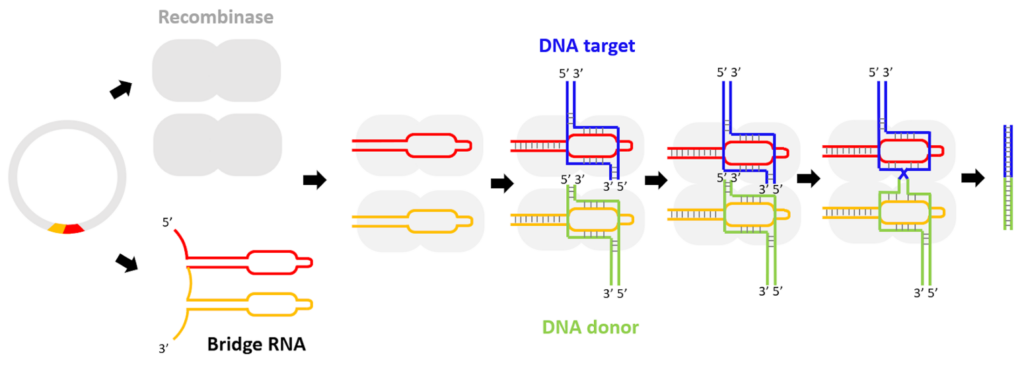
The bridge RNA technique exploits the mechanisms of mobile genetic sequences (or “jumping genes”) within the genome through cleavage and ligation, enabling modification of DNA information through insertion, excision, and inversion without exposing DNA breaks, unlike CRISPR. An enzyme called recombinase uses two loops (CRISPR uses only one) of a single bridge RNA molecule. One loop recognizes the donor sequence, which can be long, and the other loop recognizes the target DNA. Donor and target sites can be selected by designing the sequence of the bridge RNA. This new gene-editing tool was made possible by the work of the Patrick Hsu lab at the Arc Institute in Palo Alto and collaborators from the University of Tokyo.
The bridge RNA tool is based on the mechanism used by “jumping genes” such as IS110 (Insertion Sequence elements 110) that cleave and ligate themselves within and between microbial genomes. The IS110 elements consist of only a gene encoding the recombinase enzyme, flanked by DNA that joins together to produce an RNA molecule (the bridge RNA), which folds into two loops. One loop binds to the donor DNA, while the other loop binds to the target DNA where the sequence element will be inserted. The bridge RNA binds the donor and the target DNA through base-pairing interactions.
The bridge RNA technique could enable the replacement of damaged genes in cancer or inherited conditions, as well as the deletion of DNA segments that promote neurodegenerative disorders like amyotrophic lateral sclerosis (ALS) and Huntington’s disease. Overall, bridge RNA has the potential to be more revolutionary than CRISPR by enabling large-scale genome modifications.
References:
Patrick Hsu et al. Bridge RNAs direct programmable recombination of target and donor DNA, Nature (2024). DOI: 10.1038/s41586-024-07552-4. www.nature.com/articles/s41586-024-07552-4
Hiraizumi, M et al. Structural mechanism of bridge RNA-guided recombination, Nature (2024). DOI: 10.1038/s41586-024-07570-2 www.nature.com/articles/s41586-024-07570-2