Full Time Professor
Bioengineering and Chemical Engineering Department
Bacteriophages have great potential as a component expression system. Phages have been used for decades to develop new antimicrobials, monoclonal antibodies and recently to develop new biomaterials. The bacteriophage most used for this purpose is the filamentous phage M13; whose structure allows to combine the proteins of its surface with proteins of interest presented. M13 virus has the ability to infect strains of Escherichia coli strains showing F-pilus [1]. It has single-stranded DNA with a capsid composed of the PVIII protein and four smaller sheath proteins (PIII, PVI, PVII and PIX). It has 2700 copies of the PVIII protein that make up the main cylinder, and five copies of the PIII and PVI and PVII and PIX proteins at their ends. PVIII and PIII proteins are the most commonly used to perform the procedure known as bacteriophage deployment or “phage display” [1,2].
The development of antibodies depends on careful selection of antigen/antibody affinity. Usually a library of antibody variants is genetically inserted into the vector containing the viral genome where the gp3 (P III) protein from the bacteriophage cover M13 is fused to the peptide or antibody. In vitro selection allows bacteriophage filtration rounds to select those that have a high affinity for the white molecule (antigen). Selection is made by “biopanning”; an in vitro antigen-antibody binding using the ELISA method, “phage display”[1-3].
The efficient selection depends on the high affinity of the peptide (antigen) attached to the ELISA plate and the antibody fused to PIII and part of the structure of the M13 phage cover. Figure 1 shows the four-stage “biopanning” procedure. Initially, the creation of phagmanid therefore the library that allows to express variants of antibodies in the M13 virus. The second stage is the selection procedure using an ELISA, where a peptide or antigen is immobilized in the plate to which purified bacteriophages are exposed. The next washing process allows to discard those who have a low or no affinity by elution. The eluded bacteriophages are amplified in Escherichia coli, to recover the genetic material by an extraction of plasmids. Isolated plasmids (famégidos) are used to perform a next round in the process or “biopanning”. This procedure is performed 2 – 4 times to find antibodies with a high affinity [2,3].
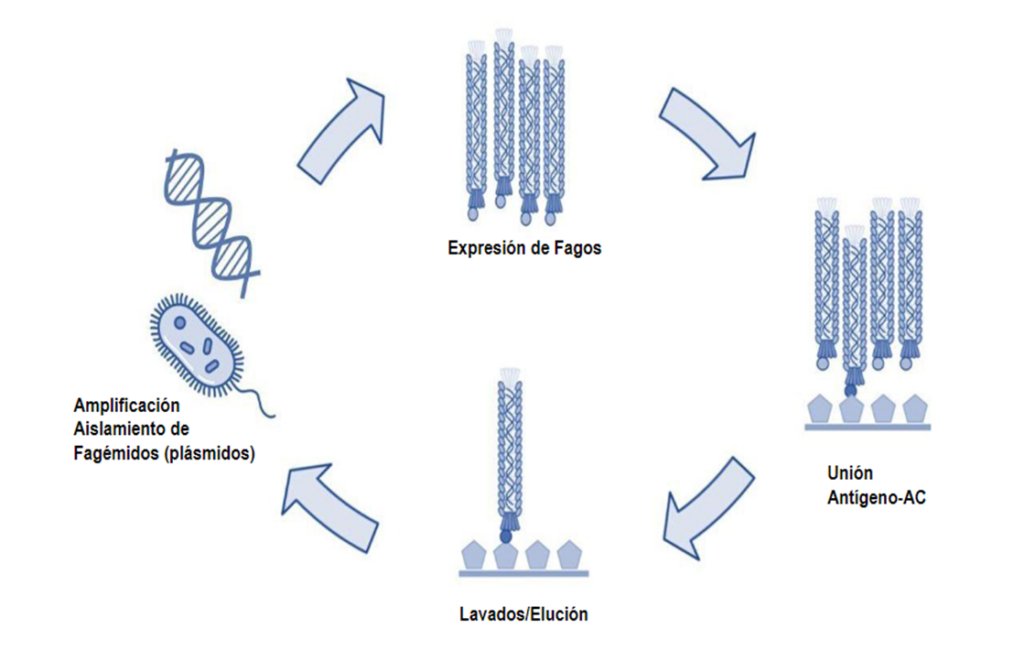
The above procedures are laborious and there is no quantitative monitoring of antigen-antibody affinity in the process. Ideally, the process of selecting new antibodies would benefit from monitoring in each round of “panning”, so as to reduce costs and concentrate efforts on molecules with high affinity and more promising [1].
It is proposed to use “molecular glue” technology to attach it to the bacteriophage deployment system. Peptides that perform biomixing in nature have the ability to covalently bind two proteins. An example is the peptides SpyTag and SpyCatcher which react spontaneously through their aspartic and lysine residues, respectively [2, 3]. Taking advantage of molecular glue, proteins can be fused as antibodies with the structure of bacteriophage M13. The proposal known as “spy-display” described in Figure 2 [3] allows the PIII protein to be fused after the library expression with the antibody mediated by SpyTag/SpyCatcher. The above offers the following advantages: A) The drawbacks of M13 phage assembly by excessive sizes of proteins fused to PIII are avoided. B) The phagoid plasmid only contains M13 proteins, improving viral production. Whereas a second expression plasmid is used for the expression of antibodies. C) Antibodies larger than single-stranded antibodies can be tested. D) The system is modular and a fluorophore, biomarker protein or enzyme could be added to monitor the antibody binding signal to decide specifically which population to use for amplification [3].

False positives are often obtained during in vitro biomolecule selection. This increases costs and reduces the efficiency of methods. In the case of antibody deployment, libraries often have truncated versions that occur during the maturation of the M13 phage in the membrane. The proposed Spy-display system allows an intact production and maturation of the M13 virus producing a “wild type” version which is post-translationally fused to the antibody library. A more robust viral population and higher infection levels are obtained. Therefore, the system shows a promising way to identify new antibodies and design variants of antibodies or synthetics that can be a contribution to testing for more effective therapies or rapid virus testing in low-cost trials [2-4].
References
1.- Passaretti, P., Khan, I., Dafforn, T.R. et al. Improvements in the production of purified M13 bacteriophage bio-nanoparticle. Sci Rep 10, 18538 (2020). https://doi.org/10.1038/s41598-020-75205-3
2.- Fierle, J.K., Abram-Saliba, J., Brioschi, M. et al. Integrating SpyCatcher/SpyTag covalent fusion technology into phage display workflows for rapid antibody discovery. Sci Rep 9, 12815 (2019). https://doi.org/10.1038/s41598-019-49233-7
3.- Kellmann SJ, Hentrich C, Putyrski M, Hanuschka H, Cavada M, Knappik A, Ylera F. SpyDisplay: A versatile phage display selection system using SpyTag/SpyCatcher technology. MAbs. 2023 Jan-Dec;15(1):2177978. doi: 10.1080/19420862.2023.2177978
4.- Qi H, Lu H, Qiu HJ, Petrenko V, Liu A. Phagemid vectors for phage display: properties, characteristics and construction. J Mol Biol. 2012;417(3):129–43. doi: 10.1016/j.jmb.2012.01.038.