Full Time Professor
Bioengineering and Chemical Engineering Department
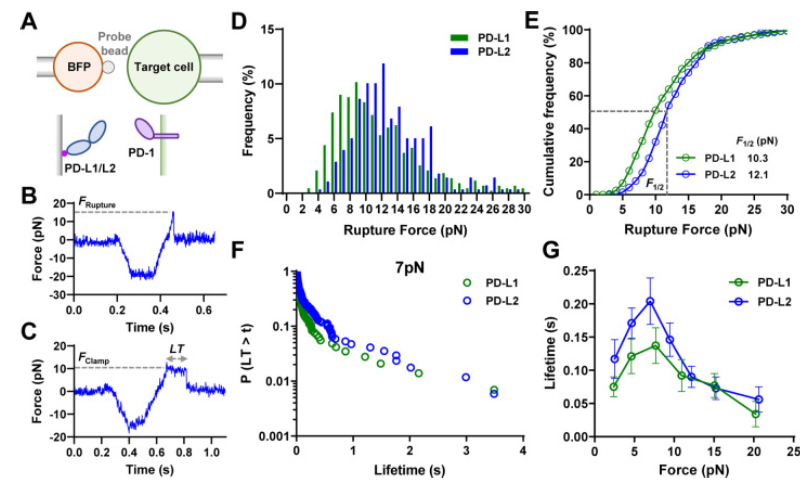
The blocking of immune control points directed to PD-1 (Programmed Cell Death Protein 1) shows great success in cancer therapy. However, the mechanism of how ligand binding (PD-L1 or PD-L2) initiates signaling to PD-1 is still unclear. As a prognostic marker for multiple cancers, soluble PD-L1 is found in patients’ serum and may bind to PD-L11, but fails to suppress the function of T cells. Previously it was shown that T cells exert endogenous forces on the PD-bonds1-PD-L2 and lead to the hypothesis that mechanical force could be critical for activation of PD-1. In the case of soluble ligand, there is no mechanical force due to the lack of mechanical support provided by ligand anchored to the surface.
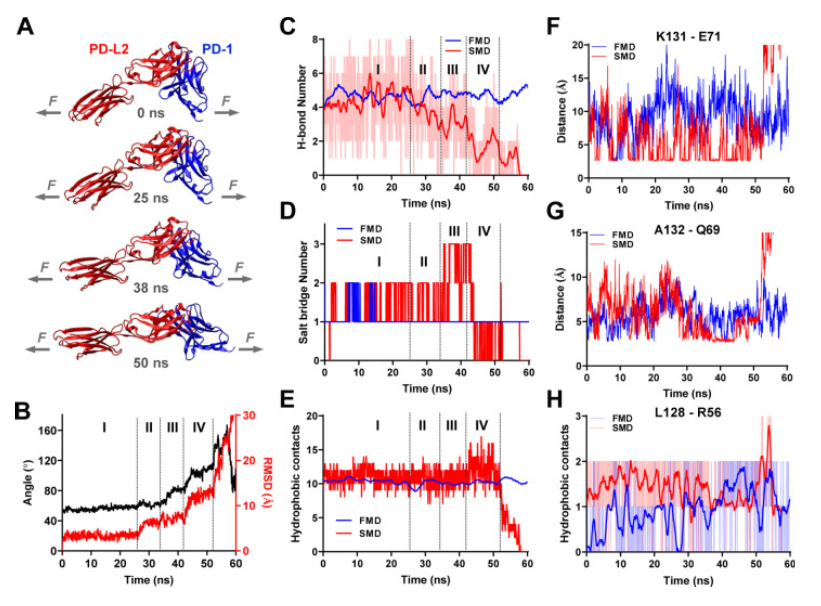
A recent study (Li K, Cardenas-Lizana, et al.) has revealed that the interaction between PD-1 and its ligands is significantly influenced by the mechanical force generated at the time of joining which is a fascinating aspect of PD-1. The study shows that the PD-1 function is eliminated or reduced when the ligand mechanical support is removed or moistened, respectively. PD-1 is a protein found in T cells, which are essential components of the immune system. It acts as an immune checkpoint that helps regulate immune responses and prevent autoimmunity. When PD-1 binds to its ligand, PD-L1, it can inhibit the activity of T cells, promoting programmed cell death in certain T cells, particularly those that are activated against specific antigens.
Catch bonds and slip bonds are molecular responses that describe two different behaviors of binding between receptors and ligands when subjected to a mechanical force. Slip bond is a bond whose useful life decreases with the increase of applied force. This is the most intuitive behavior, as a higher force usually destabilizes the links. The Catch bond is a bond whose useful life increases with applied force. This counter-intuitive behavior is essential in many biological processes, such as cell adhesion.
Key results:
Catch bonds: under low forces (less than 7 pN), PD-1 forms Catch bonds with its ligands, which means that the bond strength increases with the applied force. This extended bond life could potentially improve immune inhibitory signals.
Slip Bonds: forces greater than 8 pN transform the bonds of the PD-1 ligand into Slip Bonds, where the bond strength decreases with increasing force, This leads to faster dissociation and force-induced structural changes.
Functional implications: the ability of cells to “probe” the mechanical environment through PD-interactions1 suggests a novel mechanism for regulating immune responses based on physical signals.
Immunoengineering is an interdisciplinary field that combines principles of engineering with immunology to design innovative solutions for the treatment of diseases. By understanding the complexities of the immune system, researchers can develop therapies, Innovative diagnostics and materials to improve immune function or redirect it towards specific targets. This research opens new avenues in immunoengineering for applications of immunotherapy. Manipulating the PD-1 modulator function by changing the mechanical environment could suppress or improve PD-1 signaling. Assist in the design of new therapies by developing molecules that can interfere with force-induced changes in PD-1 and thus lead to better cancer therapies. To help in the understanding of the mechanical properties of the tumor microenvironment that could influence the function of immune cells through PD-1 interactions. Investigating in depth the force-induced structural changes in PD-1 and its ligands could provide a more complete understanding of the underlying mechanisms.
This would help in exploring the therapeutic potential of PD-1’s modular function through mechanical means, such as the use of nanoparticles or ultrasound. Finally, the role of mechanical forces in regulating PD-1 function could be investigated directly in animal cancer models. Currently UTEC’s Bioengineering professors are working on projects related to this research. If you are interested in being part of the projects you expect: Join UTEC’s Bioengineering!
References:
- Li K, Cardenas-Lizana P, Kellner AV, Yuan Z, Ahn E, Lyu J, Li Z, Salaita K, Ahmed R, Zhu C. Mechanical force regulates ligand binding and function of PD-1. bioRxiv [Preprint]. 2023 Aug 15:2023.08.13.553152. doi: 10.1101/2023.08.13.553152. PMID: 37645980; PMCID: PMC10462004.
- Zhu C., Chen W., Lou J., Rittase W. & Li K. Mechanosensing through immunoreceptors. Nature immunology 20, 1269–1278 (2019).
- Liu B., Chen W., Evavold B. D. & Zhu C. Accumulation of Dynamic Catch Bonds between TCR and Agonist Peptide-MHC Triggers T Cell Signaling. Cell 157, 357–368 (2014).
- Choi H.-K., Cong P., Ge C., Natarajan A., Liu B., Zhang Y., Li K., Rushdi M., Chen W., Lou J., Krogsgaard M. & Zhu C. Catch bond models may explain how force amplifies TCR signaling and antigen discrimination. Nature communications (2023).
- Li K., Cheng X., Tilevik A., Davis S. J. & Zhu C. In situ and in silico kinetic analyses of programmed cell death-1 (PD-1) receptor, programmed cell death ligands, and B7–1 protein interaction network. The Journal of biological chemistry 292, 6799–6809 (2017).
- Liu B., Chen W. & Zhu C. Molecular force spectroscopy on cells. Annu Rev Phys Chem 66, 427–451 (2015).
- Zhu C., Chen Y. & Ju L. A. Dynamic bonds and their roles in mechanosensing. Curr Opin Chem Biol 53, 88–97 (2019).
- Chesla S. E., Selvaraj P. & Zhu C. Measuring Two-Dimensional Receptor-Ligand Binding Kinetics by Micropipette. Biophys J 75, 1553–1572 (1998).
- Chen W., Zarnitsyna V. I., Sarangapani K. K., Huang J. & Zhu C. Measuring Receptor-Ligand Binding Kinetics on Cell Surfaces: From Adhesion Frequency to Thermal Fluctuation Methods. Cell Mol Bioeng 1, 276–288 (2008).