The discovery of gene-editing enzymes is a technological breakthrough that has an impact on various areas of knowledge, with great promise for developing efficient methods of gene therapy [1]. In this system the RNA guide (gRNA) allows a specific recognition of the cutting site in the DNA by the nucleases of the CRISPR family as the enzyme Cas9. However, the mechanisms by which, in certain circumstances, Cas9 cuts off DNA despite recognizing nonspecific noncomplementary or nonhomologous regions are unknown. Mutant versions of Cas9 have been developed that possess good nonhomologous fragment discrimination, and avoid cuts out of context. However, these versions show reduced cutting efficiency [2]. Despite numerous reports from the CRISPR system, the fundamentals of Cas9 nuclease activation when it produces nonspecific cuts in DNA out of context (“off-target”) have not yet been elucidated.
Cryo-electron microscopy (CME) provides views of the interaction between the Cas9 enzyme, the guide RNA (gRNA) and the interaction of the gRNA-DNA duplex with Cas9. CME shows interaction structures at the molecular level. For the first time, the CME technique is used to elucidate the molecular interaction kinetics of the Cas9 nuclease in its activation to perform cuts during an unusual non-homologous mating event [3].
Cas9’s HNH and RuvC domains are known to be responsible for cutting DNA. CME images show that the molecular interaction of a RuvC region is responsible for scanning the gRNA-DNA interaction to detect nonhomologous pairings. Moreover, RuvC usually stabilizes gRNA-DNA duplex events in the presence of non-homologous mating. RuvC through its “loop” domain erroneously activates the enzyme by rearranging its cut-off regions despite the existence of unmatched nucleotides in the gRNA-DNA duplex [3]. What happens if the residues in RuvC that are responsible for scanning the duplex and activating the Cas9 nuclease are mutagenized? If the mutagenesis directed by CME manages to obtain mutant versions of Cas9 that prevent an unspecific recognition without affecting its cutting efficiency would be a great advance for the use of genetic editing in a safer way.
Precision therapeutic applications hold great promise for taking advantage of the CRISPR-Cas9 system. For this methodology to be used routinely, it is necessary to minimize the risks of nonspecific cuts in the genome. Cas9 variants of “hi-fi” calls have been developed that minimize cuts out of context. In these initial versions of Cas9, the cutting efficiency or genetic editing is much lower [4].
The first step is to determine the positions of the non-paired nucleotides in the gRNA-DNA duplex. CEM images identified that when the non-paired nucleotides are at a distance of 18-20 positions from the PAM sequence, they cause a reduction rate in the DNA cut of 40 times. Nonspecific nucleotides located in positions near PAM have a lower chance of activating Cas9 in a non-homologous RNA-DNA mating event. In any of these cases, only 80% of these substrates are cut within 1 hour, when Cas9 is incubated with gRNA-DNA in vitro. Knowing that under normal conditions the in vitro cutting efficiency of Cas9 is 1 second. Thus, nonhomologous nucleotides in distal positions such as 18-20 positions with respect to the PAM sequence, usually escape surveillance and activate Cas9 causing a cut out of context. Unmanned nucleotides closer to PAM are more likely not to activate Cas9.
The cryo-microscopic structures showed that the distal end of the gRNA-DNA duplex has a relative linear conformation close to PAM, that state differs from that previously determined by the version Cas9 with homologous interaction 100% that have a typical angle of 70° (Figure 1). This means that a typical position is adopted with nonhomologous distal pairings (Figure 1, 18-20, 1 min) and is hardly detected failed mating and manages to activate Cas9. In contrast, a nonhomologous mating position close to PAM is atypical and is detected by its conformation (Figure 1, 12-14, 5 min and 60 min) and Cas9 is not activated.
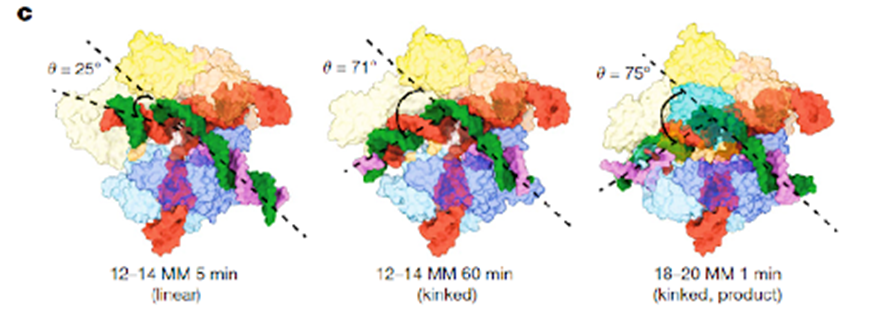
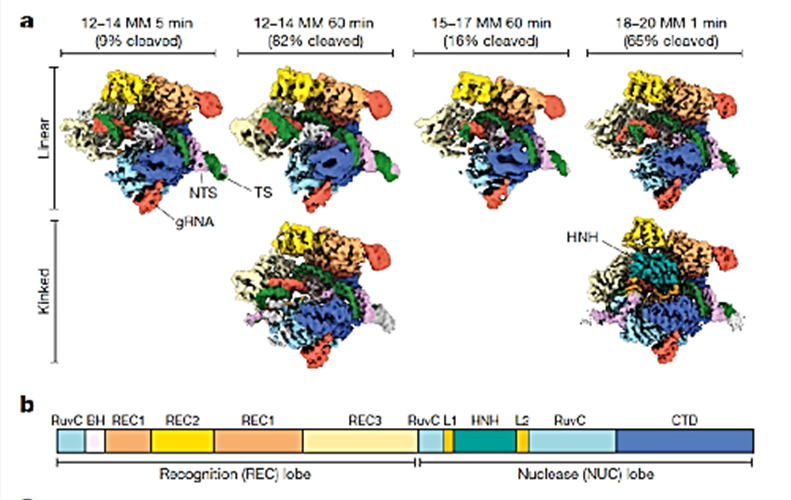
Figure 1 shows two different conformations: a linear duplex and a duplex with an angle of 70°. Cas9 conformations in both cases are identical, but the gRNA-DNA duplex distal to PAM has a path of 30 Å and is anchored with the REC3 domain of the nuclease. The hypothesis is that nonhomologous nucleotides that prevent the interaction of gRNA-distal DNA with REC3 may induce a noncanonical linear conformation rather than producing the normal doubling of 70° in the duplex, reducing the cutting efficiency. Therefore, it is postulated as a model that produces a gRNA-DNA linear duplex that precedes the 70° conformation required to activate the Cas9 cut. Nonhomologous nucleotides in positions distant from PAM (18-20), can block the conformation of 70° and escape surveillance by the REC3 domain, Cas9 is activated and the DNA is cut (Figure 2).
Like some DNA sequences that don’t have perfect homology are cut by Cas9? How do some nucleotides evade Cas9’s molecular control mechanism and not others?
After incubation of Cas9 with the DNA with nonhomologous nucleotides in the position 18-20, at the minute of incubation, 65% of the DNA has been cut. Cryo-microscopic observations of these structures allowed linear molecules and species to be identified with a normal doubling of 70° required to activate the Cas9 cut. In species with bending, the HNH domain is positioned in its active conformation. This indicates that linear molecules are part of a first event that subsequently changes to a bending that induces the anchoring of HNH and gRNA-DNA. These two stages, the linear and the doubling, allowed us to discover the role of the “linkers” regions L1 and L2 that facilitate the interaction of the HNH and RuvC cutting domains, respectively. High-precision versions of Cas9 enzymes such as “hyper-accurate-Cas9” and the high-fidelity Cas9 (Cas9-HF1) version, both with 4 prop mutations, minimize non-specific cuts, but reduce genetic editing efficiency. The mutagenized residues are located in the REC3 domain that performs active interaction with the 70°duplex. Mutants, therefore, have a lower ability to stabilize the duplex and L1 cannot so easily position HNH for cutting so editing efficiency is affected. These high-fidelity versions reduce Cas9 activation in nonhomologous nonspecific DNA, but also affect cutting at specific sites.
The R-loop model for Cas nuclease function proposes that the enzyme approximates the gRNA by creating an initial bubble in the DNA which expands to achieve gRNA mating and anchoring of the HNH and RuvC regions (Figure 3). Cryo-ME allowed the identification of amino acid residues that interact with the R-loop region, which are those that promote protein stability even in the presence of nonhomologous nucleotides as opposed to high fidelity versions, leaving the REC3 regions intact, L1 and L2 allowing to position the cutting domains HNH and RuvC. Therefore, the new SuperFi-Cas9 version has 7 mutations in the “loop” of RuvC responsible for tolerance to non-homologous sequences. This region in RuvC is thought to stabilize the duplex even in the presence of nonhomologous nucleotides by activating Cas9 to make cuts out of context.
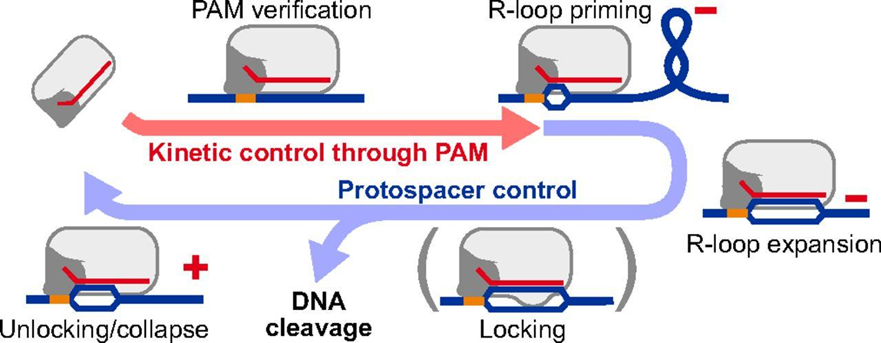
RuvC mutations in this region do not affect cutting efficiency in the case of perfect homology, but do not stabilize the duplex in case of non-homologous mating. Therefore, this version of the enzyme called SuperFi-Cas9, developed exclusively with the powerful cryo-microscope tool allows genetic editing at homologous sites and avoids unscripted cuts out of context.
Cryo-microscopy shows the potential to improve genetic editing systems that must be used in techniques of diagnosis and repair of defective genes and whose use has been limited by cuts out of context that could cause harmful effects in the cells. The CME technique will allow to obtain a new generation of “molecular scissors” improved and more reliable for their applications in Biomedicine [3,4].
References
1.- Doudna JA. The promise and challenge of therapeutic genome editing. Nature. 2020 Feb;578(7794):229-236. doi: 10.1038/s41586-020-1978-5. Epub 2020 Feb 12. PMID: 32051598; PMCID: PMC8992613. Doudna JA.
2.- Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Joung JK. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016 Jan 28;529(7587):490-5. doi: 10.1038/nature16526. Epub 2016 Jan 6. PMID: 26735016; PMCID: PMC4851738.
3.- Bravo, J.P.K., Liu, MS., Hibshman, G.N. et al. Structural basis for mismatch surveillance by CRISPR–Cas9. Nature 603, 343–347 (2022). https://doi.org/10.1038/s41586-022-04470-1
4.- Kim N, Kim HK, Lee S, Seo JH, Choi JW, Park J, Min S, Yoon S, Cho SR, Kim HH. Prediction of the sequence-specific cleavage activity of Cas9 variants. Nat Biotechnol. 2020 Nov;38(11):1328-1336. doi: 10.1038/s41587-020-0537-9. Epub 2020 Jun 8. PMID: 32514125.