Full time Professor
Bioengineering and Chemical Engineering Department
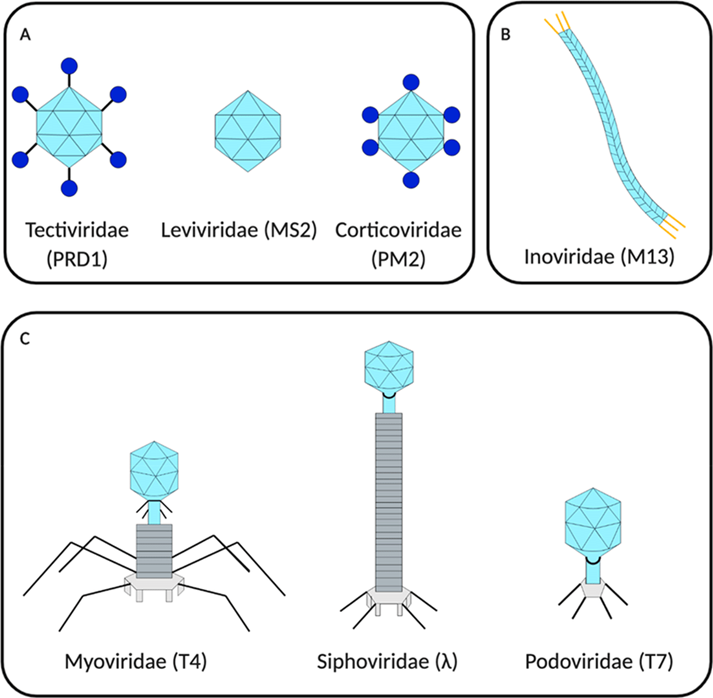
Bacteriophages are viruses that infect bacteria that can be found in all ecosystems of the planet. The infection allows them to use bacterial transcription machinery to replicate. Inside their protein capsid, bacteriophages store their genome composed of DNA or RNA with information to encode viral structural proteins. The capsid may be icosahedral (cortic virus), filamentous (phage M13) or head-tail (phage T7) (Figure 1). Bacteriophages bind to the bacterial extracellular membrane to introduce their genome into the bacterial cytoplasm. Two life cycles are known as lytic or lysogenic infection. In the lithic life cycle, bacteriophages induce rapid death of bacteria and at the same time hundreds of new viruses are released. In contrast, in the lysogenic life cycle, bacteriophages reproduce at the same time as bacteria, without destroying them [1].
Bacteriophages have become an important tool as a model of study, for understanding biology. Their study allowed us to know that DNA is the genetic material, that a codon codes for an amino acid, and to elucidate the processes of gene regulation. In the field of molecular biology, Gene cloning through DNA assembly has been possible by combining restriction enzymes that bacteria use to cut specific DNA sequences along with the process of binding DNA molecules with DNA ligase bacteriophage T4 [1, 2]. Furthermore, phage DNA polymerases have been used for sequencing DNA molecules [2]. Another growing technology for genetic editing is CRISPR-Cas (“clustered regularly interspaced short palindromic repeats”), which is based on the mechanism of defense of bacteria against bacteriophages. In applications for bioengineering, genetic modification of the genome of these viruses has allowed to modify the proteins of its surface generating new nanomaterials for the diagnosis of cancer, therapeutic agents against multidrug-resistant bacteria. Phage display (phage display) is a technology platform used in the pharmaceutical industry to evaluate antibodies, antigens and toxins related to cancer. In this system some proteins of the viral capsid are fused to peptides with affinity to eukaryotic cells, which has allowed the release of drugs to cancer cells and the development of phage-based vaccines by fusing bacteriophages with antigens from human pathogens. There are efforts to implement a vaccine with anthrax using the PA antigen delivery system to bacteriophage T4 [3, 4].
In the food industry, bacteriophages have been used as biosanitizing agents by killing pathogenic bacteria (Campylobacter jejuni and Listeria monocytogenes) or by producing enzymes (depolymerases and endolysins) to reduce the formation of biofilms on the surfaces of industrial materials. Another approach has been the application of bacteriophages as a bio-reserving agent to lengthen life in food products by killing bacteria responsible for food rot. In the dairy industry, bacteriophages can eliminate pathogens such as Staphylococcus.
In agriculture, bacteriophages have been used as agents to fight infections to plants such as Erwinia amylovora infecting apple, Ralstonia solanacearum and Xanthomonas campestri infecting tomato [5].
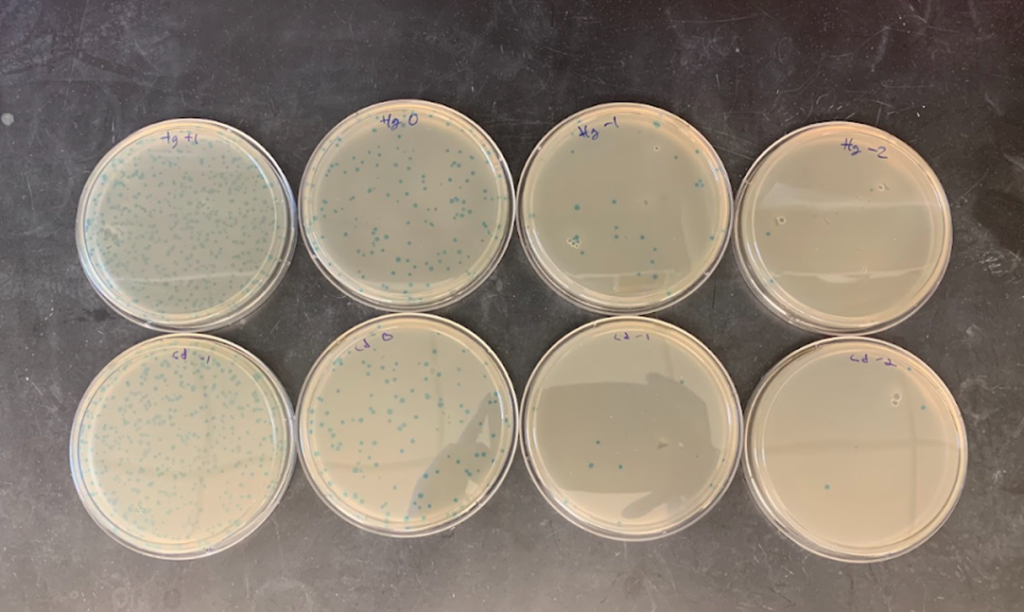
In the Department of Bioengineering and BIO Center of the UTEC, we are developing a project in collaboration with MIT (USA); for the removal of heavy metals in the environment using genetically modified bacteriophages. Ideally, it is desired to implement hydrogels capable of containing these viruses with a high capacity of absorption by heavy metals to be combined with contaminated water.
The image shows the first tests of bacteriophage construction obtained with mercury and cadmium affinity, developed by UTEC and MIT (Figure 3).
References:
[1] A. B. Monk, C. D. Rees, P. Barrow, S. Hagens, and D. R. Harper, “Bacteriophage applications: where are we now?,” Lett. Appl. Microbiol., vol. 51, no. 4, pp. 363–369, Oct. 2010, doi: 10.1111/j.1472-765X.2010.02916.x.
[2] M. Mahler, A. R. Costa, S. P. B. van Beljouw, P. C. Fineran, and S. J. J. Brouns, “Approaches for bacteriophage genome engineering,” Trends Biotechnol., vol. 41, no. 5, pp. 669–685, May 2023, doi: 10.1016/j.tibtech.2022.08.008.
[3] Tao P, Mahalingam M, Zhu J, et al. A bacteriophage T4 nanoparticle‐based dual vaccine against anthrax and plague. mBio. 2018;9(5):e01926‐e01918. 10.1128/mBio.01926-18.
[4] Hess KL, Jewell CM. Phage display as a tool for vaccine and immunotherapy development. Bioeng Transl Med. 2019 Sep 18;5(1):e10142. doi: 10.1002/btm2.10142. PMID: 31989033; PMCID: PMC6971447.[5] M. Połaska and B. Sokołowska, “Bacteriophages—a new hope or a huge problem in the food industry,” AIMS Microbiology, vol. 5, no. 4, p. 324, 2019, doi: 10.3934/microbiol.2019.4.324.